All of MIPLab and collaborators’ codes and developed softwares. We also maintain a GitHub repository associated to the lab.
In-House Software
iCAPs Toolbox
FMRI deconvolution using state-of-the-art sparsity-driven regularization
TbCaps
ToolBox for Co-Activation Patterns
PLS implementation
Matlab algorithm for Partial Least Squares (PLS)
Graph Slepians
The design of bandlimited graph signals that maximize energy in a subgraph as the graph equivalent of Slepians is used to generalize Laplacian embedding and graph clustering.
Dynamic functional connectivity
Matlab code for determining eigenconnectivities from dynamic functional connectivity measures

OpenNFT
An open-source Python/Matlab framework for real-time fMRI neurofeedback training

Ceci n’est pas un cerveau
3D-printable Shepp-Logan phantom with analytical expressions in k-space
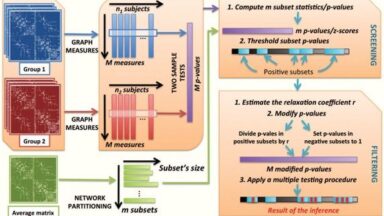
Statistical Parametric Connectome (SPC)
Two-step procedure improves sensitivity of statistical testing connectomes

Functional connectivity pipeline
Matlab pipeline for wavelet-based functional connectivity analysis
Surrogate Graph Signals
Generate surrogate data for signals defined a graph backbone
Wavelet-Based Statistical Parametric Mapping (WSPM)
Toolbox for SPM to perform statistical testing using the wavelet transform and without Gaussian smoothing
Non-local means (NLM)
Extensions for NLM such as Stein’s unbiased risk estimate (SURE) and optimal linear expansions of multiple NLMs
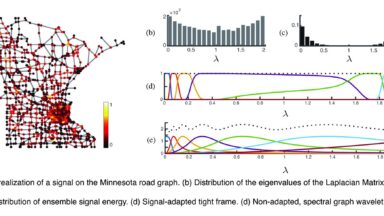
Graph signal decompositions
Spectral decompositions designed for graphs
Riesz steerable pyramid
Steerable wavelet transform for advanced image analysis such as rotation-covariant texture learning